Abstract
Background: Myeloid sarcoma (MS), or granulocytic sarcoma, is a rare extramedullary tumor of immature myeloid cells. In light of its distinct biologic behavior compared to that of conventional AML, namely, homing and clustering outside the hematopoietic system, being responsive to immune checkpoint inhibitors while refractory to common myeloid toxins, we hypothesized that MS could share some of the genetic abnormalities commonly found in solid tumors demonstrating features that mimicking them. The aim of this study was to explore this using a more expanded panel of cancer genes, which are not necessarily restricted to known AML-associated genes, to gain insight into the molecular pathogenesis of MS and to identify potential drug targets.
Methods: We retrospectively identified and collected clinical data of 62 patients with a diagnosis of MS made between March 2003 and May 2016 at Seoul National University Hospital (SNUH). Of these, 13 patients went through planned panel sequencing of 83 genes using formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks. The targeted genes were concentrated more on well-known oncogenes than on relatively unknown genes, and include detection of single nucleotide variants (SNVs), insertion/deletion (indels), copy number variations (CNVs), and gene fusions.
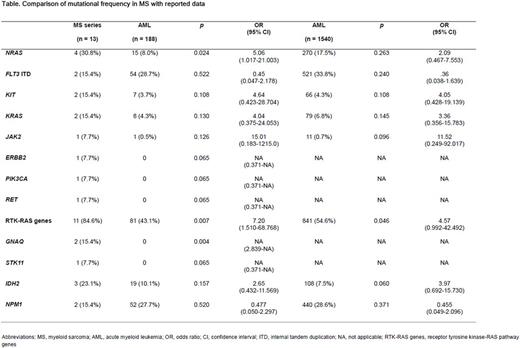
Results: All 13 sequenced cases had at least one well-known oncogenic mutation, and more than one mutation was found in four patients, with all cases positive for the IDH2 and/or NPM1 mutation in the current study exhibiting another co-mutation. Strikingly, most of them (11 out of 13 cases) had a mutation in the genes of the receptor tyrosine kinase (RTK)-RAS pathway. NRAS was the most frequent genetic alterations among these, affecting four cases. FLT3 ITD, KIT, and KRAS eachwere found in 2 patients, whereas ERBB2, JAK2, PIK3CA, and RET each were identified in 1 case. When compared with that from the reported data of AML, their mutational frequency as a group was 84.6%, which was significantly greater than that of 43.1% and 54.6% in AML, as reported from whole genome and whole exome sequencing in the Cancer Genome Atlas and extensive target sequencing involving more than 1500 AML patients, respectively (p= .007 and p= .046, respectively).
Conclusion: In conclusion, the pattern of molecular derangements in MS was generally consistent with that in AML, but MS was apparently more enriched with mutations of the RTK-RAS pathway genes, sharing genetic commonalities with solid tumors than with AML. Future studies are warranted to elucidate their therapeutic and prognostic implications, as biochemical inhibition of oncogenic Ras signaling is being actively studied, as well as the detailed molecular mechanism underlying their distinct phenotypic expression.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal